Example Target Analysis
This directory contains a example analysis of Hyb-format data, published in the quick Crosslinking and Sequencing of Hybrids (qCLASH) experiment described in: Gay, Lauren A., et al. "Modified cross-linking, ligation, and sequencing of hybrids (qCLASH) identifies Kaposi's Sarcoma-associated herpesvirus microRNA targets in endothelial cells." Journal of virology 92.8 (2018): e02138-17.
- The analysis is carried out in multiple example implementations which produce identical output:
via the Command-Line
via the Python3 API
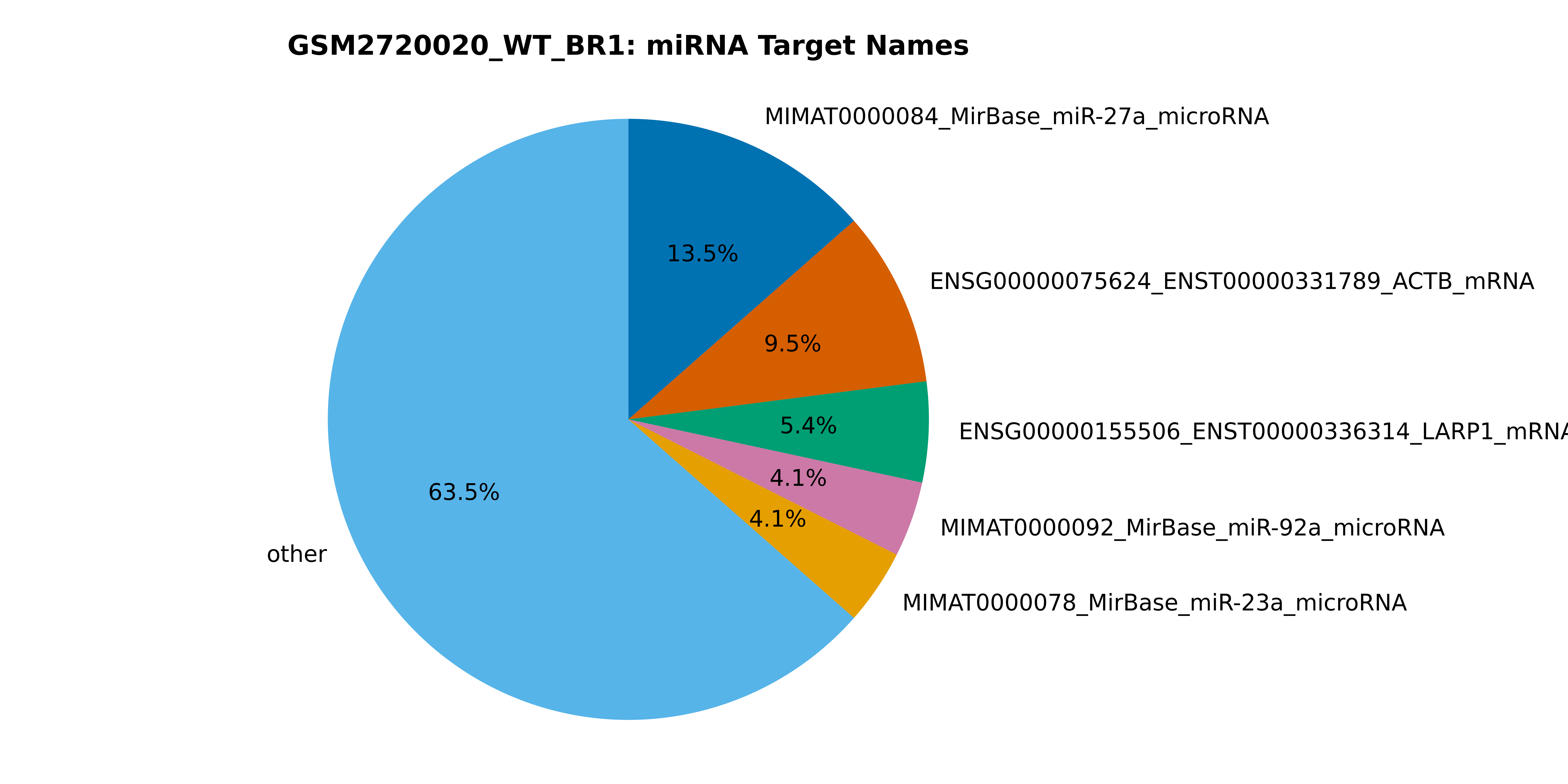
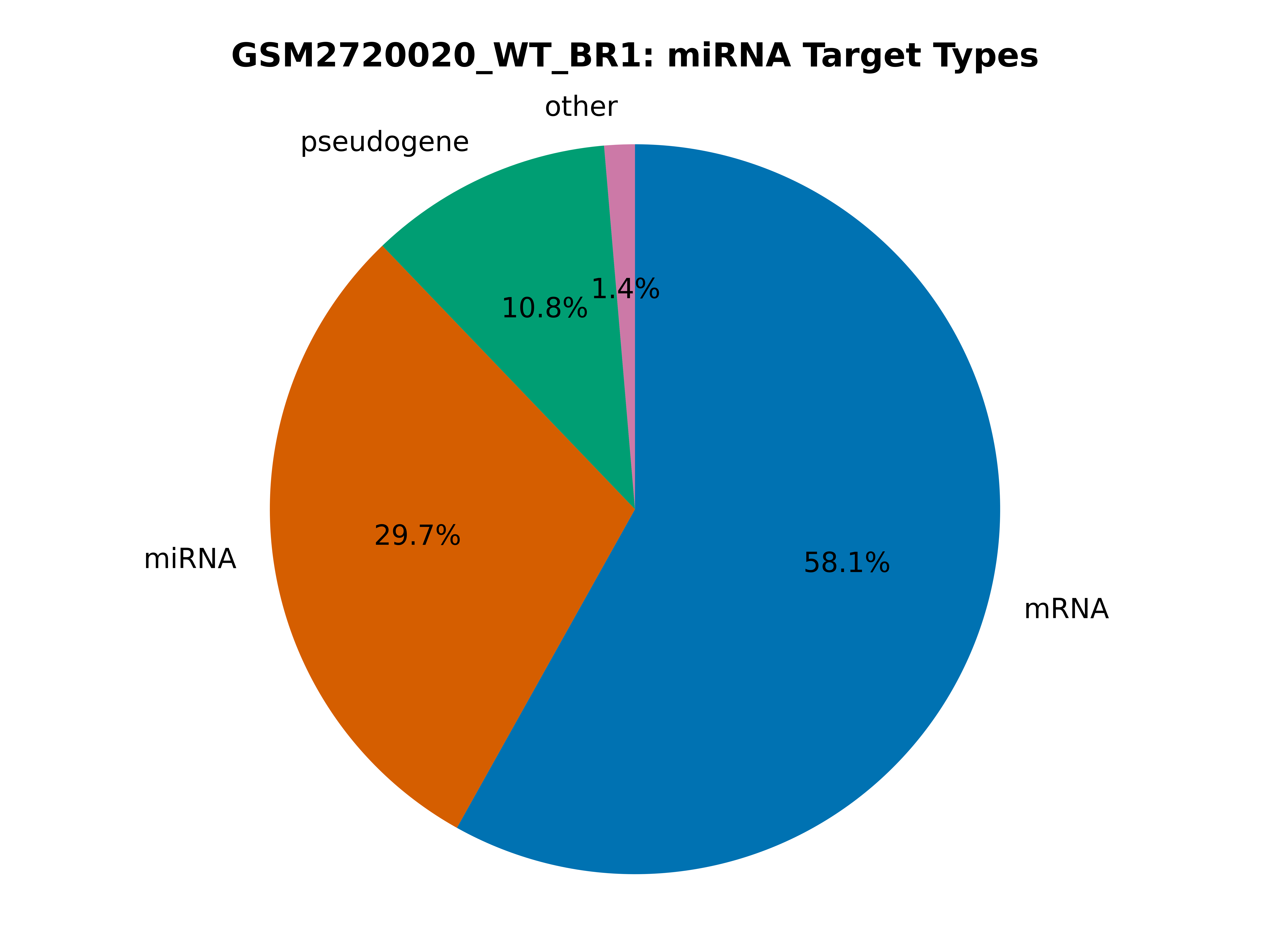
This analysis specifically filters and analyzes the kshv-miR-K12-5 miRNA arising from Kaposi's Sarcoma-Associated Herpesvirus (KSHV), which has the assigned type "KSHV-miRNA". Both individual and summary output files are produced.
Hybrid sequences generated by the Hyb program are available at NCBI Gene Expression Omnibus (GEO) GSE101978, at:
The data files can be downloaded and uncompressed by using the command:
$ sh ./download_data.sh
The unpacked hyb data-file require ~130 MB of space. The completed output of the analysis requires ~20 MB of space.
Target Analysis Example Output